Key Takeaways:
- Google and Yale’s C2S-Scale 27B model generated a new hypothesis about cancer cell behavior, later confirmed through lab experiments in living cells.
- The model highlighted silmitasertib, a kinase inhibitor, as a conditional amplifier of immune response when combined with low-dose interferon.
- Lab tests showed a nearly 50% increase in antigen presentation, helping previously unresponsive tumors become more detectable by the immune system.
- Only the 27B-parameter version of the model demonstrated the reasoning needed for the discovery, underscoring the potential of scaled AI in biological research.
- Google’s DeepMind has laid a strong foundation in biomedical AI with models like AlphaFold, AlphaMissense, and AlphaGenome, each advancing our understanding of how genetic mutations influence health.
- What began with predicting protein shapes has evolved into a broader mission to decode the entire genome, showing that C2S Scale 27B is not an isolated achievement but part of a long-term shift in using AI to uncover the biological roots of disease.
Table of Contents
A Model Built to Understand the Language of Cells
A major breakthrough in cancer research may be on the horizon, thanks to a new AI model developed by Google in collaboration with Yale University. The model, called C2S-Scale 27B, generated a novel hypothesis about cancer cell behavior which experimental research in living cells has since confirmed, laying the groundwork for future therapies designed to make tumors more visible to the immune system.
The newly launched foundation model, C2S-Scale 27B, is based on Google’s Gemma open models and is tailored to decode the complex language of individual cells. With 27 billion parameters, the model was designed to handle the intricacies of single-cell biology, particularly in cancer research.
One of the key challenges in cancer immunotherapy is that many tumors remain “cold” — they do not alert the immune system to their presence. To address this, the model was tasked with identifying a drug that could amplify immune signals only in environments where low levels of interferon, a natural immune-signaling protein, are already present but insufficient on their own to trigger a response.
Using a virtual screening process across more than 4,000 drug candidates, C2S-Scale 27B pinpointed silmitasertib, a kinase CK2 inhibitor, as a standout. The model predicted that the drug would significantly enhance antigen presentation — a process that helps the immune system detect cancer — but only in the right immune context. Importantly, this prediction had not been documented in existing literature.
Prediction Confirmed: Silmitasertib Shows Strong Results in Lab Tests
Researchers put the AI’s hypothesis to the test in laboratory experiments using human neuroendocrine cells, which the model had not previously encountered. The results were striking. While silmitasertib or low-dose interferon alone produced little change, their combination led to a nearly 50% increase in antigen presentation. This suggests a synergistic effect that could help turn “cold” tumors “hot,” making them more visible to the immune system and potentially more responsive to treatment.
The team at Yale is now investigating the mechanism behind the silmitasertib effect and is testing other predictions generated by the model in different immune scenarios.
Power in Scale: How Larger AI Models Enable Smarter Discoveries
The research builds on earlier work from Google demonstrating that biological models improve with scale, similar to language models — the larger the model, the more advanced its capabilities. In this case, only the 27-billion-parameter version of C2S-Scale was able to perform the complex, context-dependent reasoning required for this discovery.
While still in the early stages, this experiment provides a compelling proof of concept for using large-scale AI models in drug discovery and immunotherapy. With further preclinical and clinical testing, the pathway uncovered by C2S-Scale 27B could one day inform real-world cancer treatments.
Part of a Larger Shift: Google’s DeepMind Was Ahead of the Curve
While C2S Scale 27B may seem like a leap in cancer research, it is actually part of a steady and evolving journey within Google’s AI labs that has been quietly transforming the field of genetics for years.
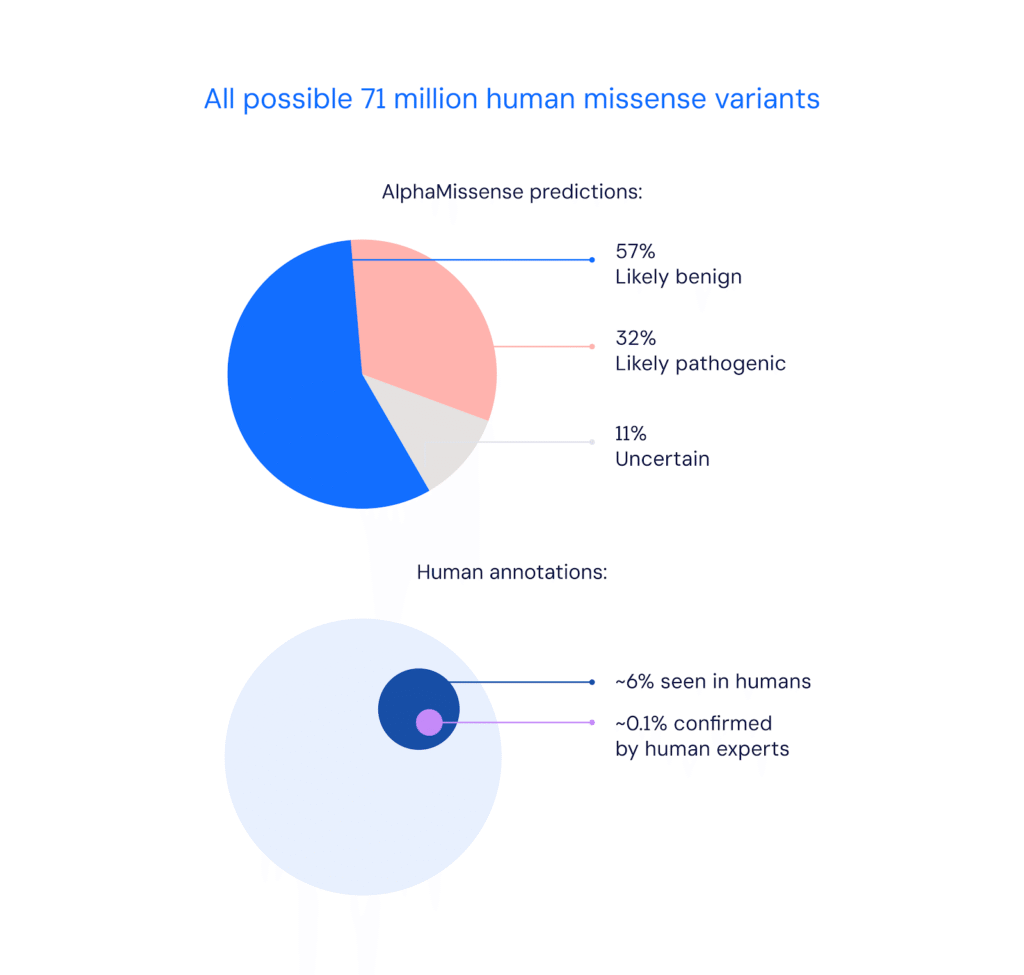
Back in 2023, Google’s DeepMind introduced AlphaMissense, a model designed to predict whether single-letter genetic mutations, known as missense mutations, are likely to cause disease.
These mutations, though often subtle, can have a profound effect on protein function and are associated with a wide range of disorders, from cancer to cystic fibrosis.
AlphaMissense tackled a daunting problem in genomics, which is the sheer volume of potential mutations, most of which had never been studied. This meant that traditional methods could confidently classify only a tiny fraction of them.
In contrast, AlphaMissense analyzed over 71 million possible variants and was able to categorize nearly 89% as either likely benign or pathogenic. Its predictions were later validated through lab testing, and the results were made publicly available to support global research efforts.
This work built on DeepMind’s earlier success with AlphaFold, the AI system that accurately predicts protein structures from amino acid sequences. While AlphaFold focused on form, the three-dimensional shape of proteins, AlphaMissense shifted the focus to function, estimating the potential impact of mutations in protein-coding regions.
Then, in 2025, DeepMind released AlphaGenome, a model that pushed this line of research even further.
While AlphaMissense examined mutations in regions that directly code for proteins, AlphaGenome turned its attention to the vast non-coding regions of DNA, areas that do not produce proteins but play critical roles in regulating gene expression.
More specifically, AlphaGenome was designed to interpret how mutations in these sequences might disrupt normal biological processes or contribute to disease, offering new insights into areas that traditional tools often failed to analyze.
Together, these models reflect DeepMind’s expanding ambition: to build AI systems that not only describe biology, but help decode and anticipate its most complex mechanisms, from the folding of proteins to the genomic signals that drive disease.
Read More: Your Job Is Safe from AI, For Now. But the Clock Is Ticking, Yale Researchers Warn







